Ecosystem networks and flux
 Oct 05, 2016 • 7:52 PM UTC
Oct 05, 2016 • 7:52 PM UTC Unknown Location
Unknown Location 140x Magnification
140x Magnification Microorganisms
Microorganisms
laksiyer
Human observer of life. https://sukshmadarshin.wordpress.com
97posts
1255comments
5locations

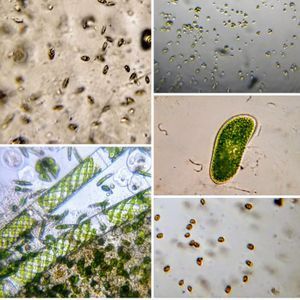
Any species that we observe under a microscope is part of an ecological network that depends on resources and interactions with other species. Think of all the ecosystems we are so used to seeing and reading about regularly: Serengeti, Yosemite, Kaziranga, Madagascar and Artic, each of these have completely different profiles of macro-species from the other. Relationships between species can be complex and includes: symbiosis, cooperation, competition and predator-prey. Can we pry out these relationships when we observe different species under a foldscope. can we distinguish these from chance co-occurrences?
Exactly about a year ago, I started sampling life in this pond near my place. I sampled water from the same spot each time to see how many species I could find. For most of the year, the pond would be covered with duckweed and the samples in bottles would last stably for months (see posts Day 1 , Day 6 , Day 17 , Day 27 , Day 80 from a previous sampling). Several species were observed at a boring regularity. A month back, I examined the pond and to my surprise there was no duckweed. A couple of weeks ago, I took a sample of pond water and in one week the water was smelling like rotten eggs. Clearly there has been a change in the ecosystem, and this suspicion was corroborated by the kinds of species I saw, most of which I hadn’t seen previously.
Note: Several videos follow. One thing that is really fun to see is the kinds of motion of the different organisms. I have also embedded my comments in the videos to reduce text.
Ciliates
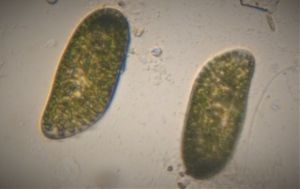
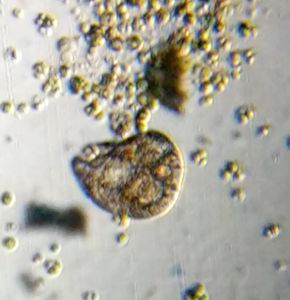
1. Video 1. Paramecium . The more I see this ciliate, more questions I have. See the video for what was running in my head. In the last part of the video, I included a 600x compound microscope view. since I wanted to show the spiral bacteria trapped in the food vacuole.
Exactly about a year ago, I started sampling life in this pond near my place. I sampled water from the same spot each time to see how many species I could find. For most of the year, the pond would be covered with duckweed and the samples in bottles would last stably for months (see posts Day 1 , Day 6 , Day 17 , Day 27 , Day 80 from a previous sampling). Several species were observed at a boring regularity. A month back, I examined the pond and to my surprise there was no duckweed. A couple of weeks ago, I took a sample of pond water and in one week the water was smelling like rotten eggs. Clearly there has been a change in the ecosystem, and this suspicion was corroborated by the kinds of species I saw, most of which I hadn’t seen previously.
Note: Several videos follow. One thing that is really fun to see is the kinds of motion of the different organisms. I have also embedded my comments in the videos to reduce text.
Ciliates
1. Video 1. Paramecium . The more I see this ciliate, more questions I have. See the video for what was running in my head. In the last part of the video, I included a 600x compound microscope view. since I wanted to show the spiral bacteria trapped in the food vacuole.
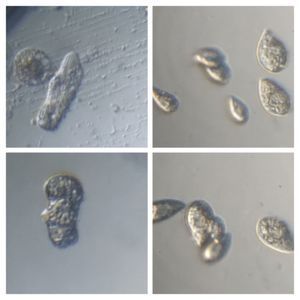
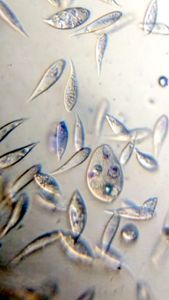
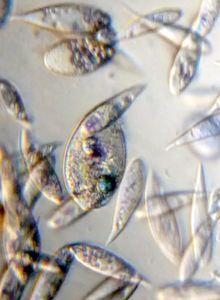
2. Video 2. Ciliate-19176-2-LI. A very Paramecium -like ciliate, although differs from it in many ways.
3. Video 3. Ciliate-19176-3-LI. An Oxytricha -like ciliate with cilia/cirri under the oral apparatus. Black granules are seen in the cytoplasm.
4. Video 4. Ciliate-19176-4-LI. My best guess is this is related to Epalxis .
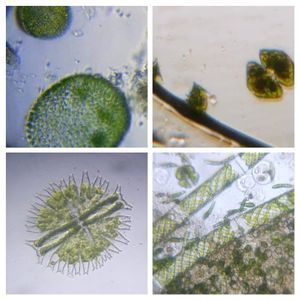
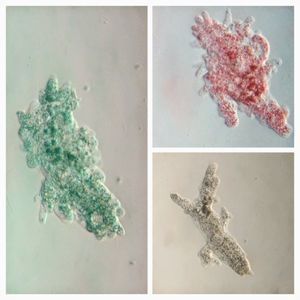
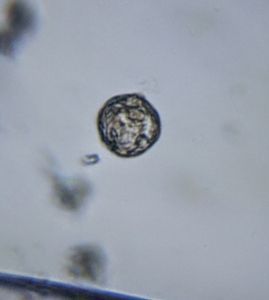
Diatoms . At least 5 types of Diatoms were seen in the sample.
5. Diatom-19176-5-L1. My best guess is Pinnularia . One can see the nice gliding motion. Those droplet like structure in the diatom might be oil droplets.
5. Diatom-19176-5-L1. My best guess is Pinnularia . One can see the nice gliding motion. Those droplet like structure in the diatom might be oil droplets.
6-9. Other diatoms

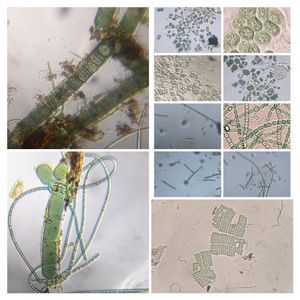
Diatom-19176-6-LI: Fragilriopsis-like, Diatom-19176-7-LI: Amphipleura, Diatom-19176-6-LI: Square-shaped diatom, Diatom-19176-6-LI: Navicula? Euglenozoans . These are basal eukaryote related to the more deadly kinetoplastids ( Leishmania, Trypanosoma ). I particularly like them as they have a unique modified DNA base called Base J and some of my own work has been intimately involved in the discovery of an enzyme involved in this modification.
10. Video of Euglena . Thank you Dr. John Hall of University of Maryland for the identification. No amount of computational knowledge of Euglena prepares you for the first time view.
10. Video of Euglena . Thank you Dr. John Hall of University of Maryland for the identification. No amount of computational knowledge of Euglena prepares you for the first time view.
11. Leaf-like Phacus , wonderful motion pattern.
12. An unknown Euglenozoan with a very interesting gyrating motion pattern. Euglenoid-19176-12-LI
13. Another unknown Euglenozoan with a Phacus like moving pattern. Euglenoid-19176-13-LI
14. A flagellate with a wiggly motion. Flagellate-19176-14-LI
15. A chlorophyte. Chlorophyte-19176-16-LI. While this could be a diatom shell, I still think it looks more like a charophyte type chlorophyte.

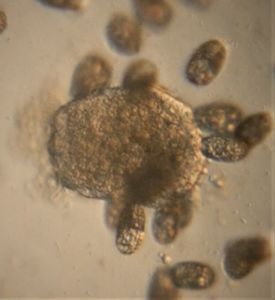
17. Amoeboflagellate (Amoeboflagellate-19176-17-LI ) : These transition between the flagellate and amoeboid forms. Although I did not see the transition, I have a strong suspicion based on previous observations that they belong to this group. Watch the video for a particular pattern of arrangement. Is this a chance observation or does it have a deeper signficance.
18. Nematodes (round worms). Nematode-19176-18-LI
The longest nematode I have seen. Many nematodes attack plant roots. I wonder if this was the cause for the death of the duckweed?
The longest nematode I have seen. Many nematodes attack plant roots. I wonder if this was the cause for the death of the duckweed?
19/20. Bacteria : Several different types of bacteria were seen, but two that caught my attention were this Beggiatoa like long bacterium (Bacterium-19176-19-LI) and these long Spirllum like spiral bacterium (Bacterium-19176-20-LI). In Saad’s post on
Paramecium , we see this long Beggiatoa -like bacterium along with Paramecium . Just like in the video below.
Paramecium , we see this long Beggiatoa -like bacterium along with Paramecium . Just like in the video below.
Now in this pond water derived system all the species were observed in the same cross-section of space. Some might be chance associations, others might have various levels of dependencies on the state of resources in the environments and other species. Over multiple such observations could we now create ecological networks based on co-occurrence of species, where the nodes are the species and the edges are co-occurrences. The thickness of the edges is proportional to the number of co-occurrences. For example, if I see Paramecium and the long chained Beggiatoa like bacterium this will have a co-occurrence score of 1. Now if I see it again (as I think I do in Saad’s post), then it will have a co-occurrence score of 2 (a thicker edge). Over multiple observations, we will start noticing the strongest associations and eliminate the ones observed by chance. Thus we could build an ecological network. A very basic (and rather unaesthetic) version is below (Click on image for high-res). Its an all-vs-all network, note the thicker Paramecium-bacterium edge.
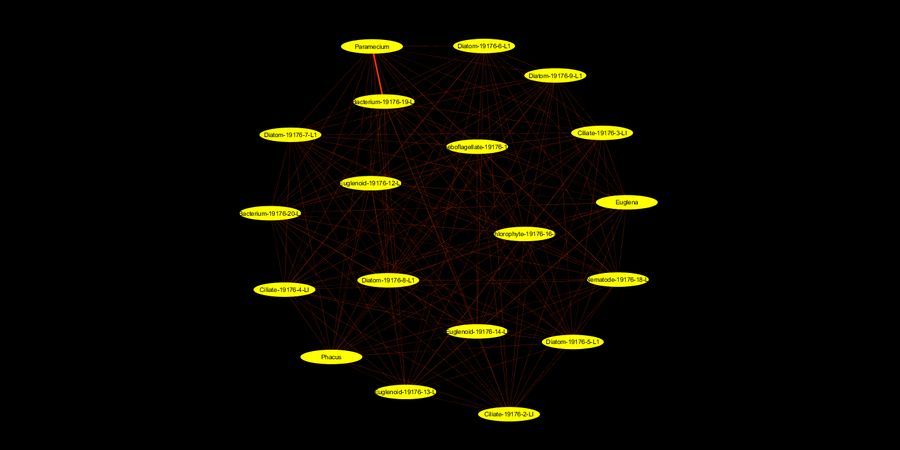
P.S. My aquarium lasted more than a week on this occasion. Paramecium was still seen after 3 days and Oxytricha after a week.
This post is open to read and review on The Winnower.
This post is open to read and review on The Winnower.
Sign in to commentNobody has commented yet... Share your thoughts with the author and start the discussion!

 0 Applause
0 Applause 0 Comments
0 Comments_300x300.jpeg)