Exploring a Slime Mold-2: Tracks outside and within
 Feb 11, 2018 • 7:44 PM UTC
Feb 11, 2018 • 7:44 PM UTC Unknown Location
Unknown Location 140x Magnification
140x Magnification Microorganisms
Microorganisms
Laks Iyer
Human observer of life. https://sukshmadarshin.wordpress.com
97posts
1255comments
5locations

View in Media Gallery
Continuing from my previous post , you can imagine that all my free time is spent trying to do as much with the slime mold. By now I am quite certain that this is some kind of Fulgio species (perhaps Fulgio septica ). However,one needs to really look for several traits before deciding the species, and so I will have to study this a bit more. The plasmodium stage of the myxomycete is fascinating and very amenable to time lapse videography. A study of these videos taken from several different plates provide an idea of the foraging strategy of the myxomycete plasmodium. There appear to be at least 3 stages: 1) a jiggly stage where the plasmodium pulsates in one place. 2) A spreading stage where the leading edge spread out “legs” in various directions and an 3) optimizing stage which thins out regions that it has already explored to often form vein-like structures. How would you describe this? Could you think of a mathematical model that fits this kind of foraging? Perhaps it has to deal with how to spread out a fixed volume of material, first a random lunge in various directions and then optimization.
Subculturing of the plasmodium is super easy. Transfer an oat that has been taken over by the slime mold to another plate with moist filter paper and add more oats. You can even do food choice experiments, like the one shown below.

View in Media Gallery
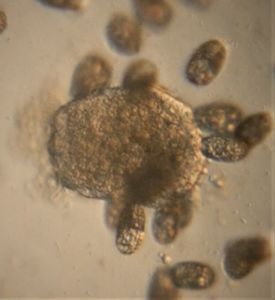
In this I wanted to see if the myxomycete plasmodium at the center in 1 chooses to go to the barks of wood, oats or yeast pellets. But when you view its progression from 1-5, you will see that it processes them serially, first to the wood bark, then the the yeast, then to one of the oats and then to the other bark. I had two burning questions after these experiments. 1) How might this appear under a microscope? 2) I observed dark tracks laid down wherever the plasmodium moved and formed vein-like structures. In the picture above, in pictures 3, 4 and 5 you can see the dark tracks that are on the path of the myxomycete. Could these dark tracks be some kind of flags, an equivalent of spatial memory, that lets a plasmodium know that this place has been visited? After 2 months of experiments, I have never seen the plasmodium go back on its tracks!! What is the source of these tracks–another pigment or perhaps the food itself?
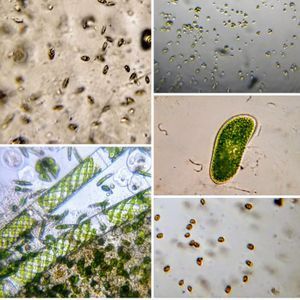
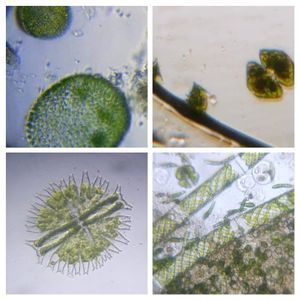
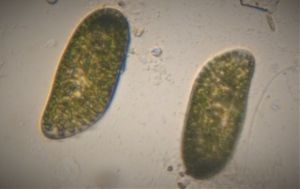
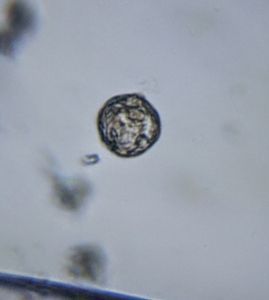
To answer these, I turned to the foldscope and it took me a few trials to understand how to get a good myxomycete sample for live viewing. At first I was a bit disappointed when I foldscoped a scraping of the plasmodium– it was not very revealing as the pigment causes the plasmodium to be opaque, but the good thing was that one could see dark spots that corresponded to the black marks on the tracks, suggesting that these were exported out from the cell and laid out along the path of the plasmodum. The source of the black pigment is still unclear. Could it be from the bark? or is it a hyperchromic yellow pigment or an entirely different pigment? There are also vacuoles and perhaps pigment containing vesicles that are seen in the videos below.
To answer these, I turned to the foldscope and it took me a few trials to understand how to get a good myxomycete sample for live viewing. At first I was a bit disappointed when I foldscoped a scraping of the plasmodium– it was not very revealing as the pigment causes the plasmodium to be opaque, but the good thing was that one could see dark spots that corresponded to the black marks on the tracks, suggesting that these were exported out from the cell and laid out along the path of the plasmodum. The source of the black pigment is still unclear. Could it be from the bark? or is it a hyperchromic yellow pigment or an entirely different pigment? There are also vacuoles and perhaps pigment containing vesicles that are seen in the videos below.
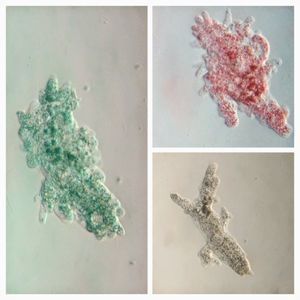
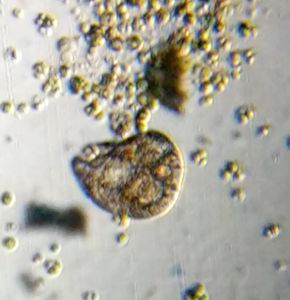
After several trials, I realized that when I took a piece of plasmodium growing on the filter paper and left it on a slide within a chamber, with a little bit of water in it to keep it moist, it would grow out of the paper after half a day and forage on the rest of the slide (see below picture). By now the original sample had starting growing on the plate bottom (see picture below), so I directly took some of the slime moldand put in the slide chamber(see pictures below). Slides like these last 2-4 days without drying out and make for good microscopic study. This could also be used for studying phagocytosis (but that is for another post).

View in Media Gallery
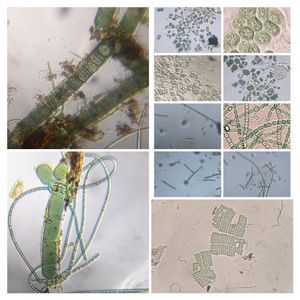
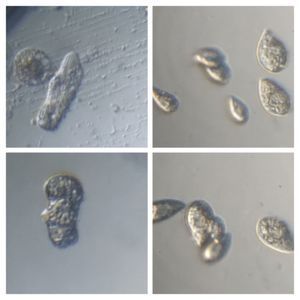
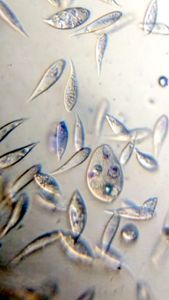
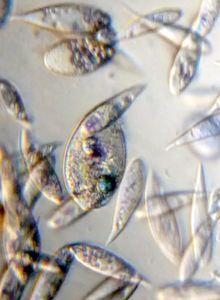
And Aha!!! I finally got to see how the plasmodium streams the cytoplasm during foraging. It is bidirectional and quite vigorous. It almost appears like there is a pump that pulls and pushes cytoplasmic material through a giant cell.Further, the membrane buckles along various fronts just like an amoeba’s cell. The following collation of videos is split into two parts. In the first, are 3 time lapses of the myxomycete plasmodium. In the second are cytoplasmic streaming videos. One question that comes to mind is how does the myxomycete efficiently adjust its internal highways by this apparent “pumping-like” pulsating action? Is there a nerve center or is this a distributed process? Could we learn something from this, such as optimizing traffic of various types, such as the internet or roadways/railways? Let me know what you think.
Second video follows
So lets step back and think through this? This is a huge cell with multiple nuclei. While it deceptively looks like a river flowing, the cytoplasm is hardly fluid. What is it that is streaming inside, and how is the speed controlled? What is the basis of movement? Studies have shown that amoeboid motion involves proteins that also go into the machinery involved in muscle contraction, i.e.actin-myosin tracks (see references 1,2 , 3 ). Further, transport of cellular material involves the vesicular transport system, i,e, along microtubule tracks crisscrossing the cell, with GTPases powering vesicles along microtubule track. So an extensive track network exists within the cell. One would think that if you peer into the genomes of these organisms they might have something special that distinguishes them from other organisms.
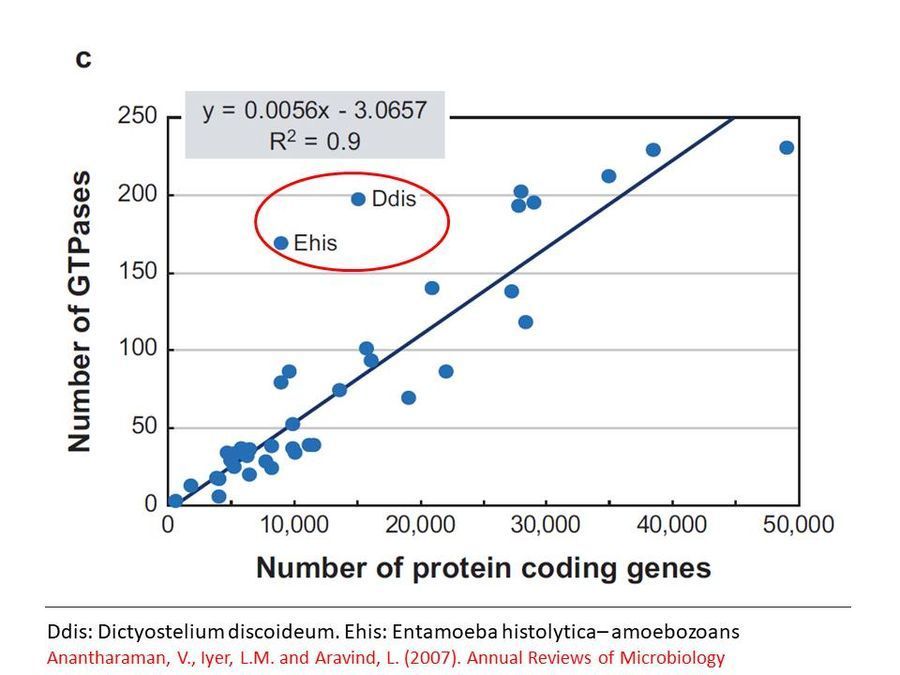
Genome analyses show that the GTPases involved in vesicular transport are present in higher than normal numbers per genome in slime molds and amoebae as seen in this graph below ( Click here to access reference ) . This might reflect the adaptation of these organisms to their unusual life style. Clearly while Dictyostelium has been well studied in many of these molecular aspects, one can only wonder what the molecules might be in this slime mold. Perhaps some smart molecular biologist will take this further.
Genome analyses show that the GTPases involved in vesicular transport are present in higher than normal numbers per genome in slime molds and amoebae as seen in this graph below ( Click here to access reference ) . This might reflect the adaptation of these organisms to their unusual life style. Clearly while Dictyostelium has been well studied in many of these molecular aspects, one can only wonder what the molecules might be in this slime mold. Perhaps some smart molecular biologist will take this further.
View in Media Gallery
Finally, I played a lot with staining the plasmodium, but that is for another day.
Special thanks to Dr. Steve Stephenson of the University of Arkansas for his wonderful book and also for regularly replying to my questions through email.
Special thanks to Dr. Steve Stephenson of the University of Arkansas for his wonderful book and also for regularly replying to my questions through email.
Sign in to commentNobody has commented yet... Share your thoughts with the author and start the discussion!

 0 Applause
0 Applause 0 Comments
0 Comments_300x300.jpeg)